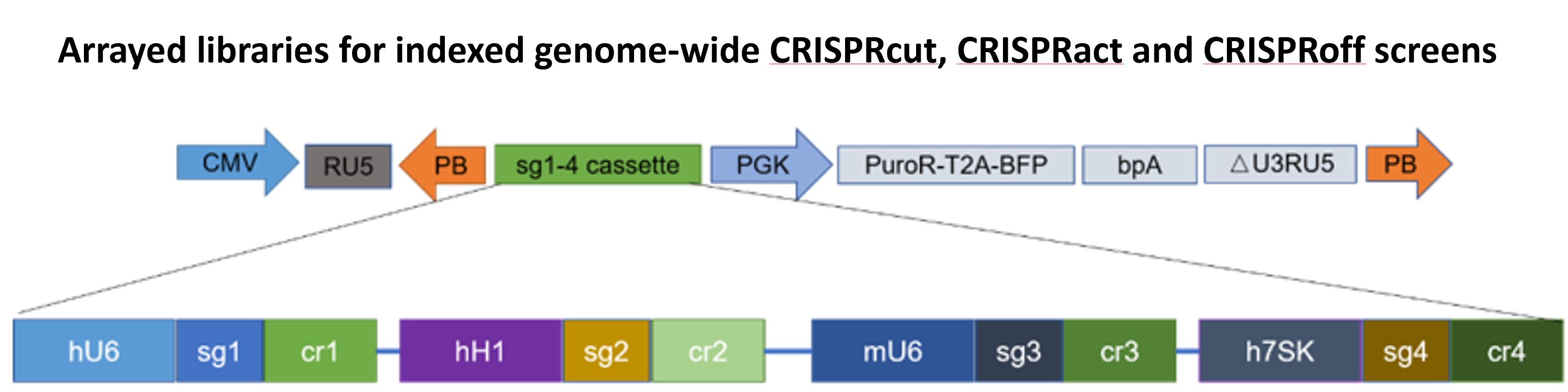
Next-generation arrayed CRISPR libraries enable biochemical, morphological, and non-autonomous phenotypic screens that pooled libraries cannot handle.

Phenotypic CRISPR screens are instrumental many fundamental biological phenomena, and arrayed CRISPR libraries extend the screening territory to cell-nonautonomous, biochemical and morphological phenotypes. We have recently completed the generation of two human genome wide arrayed plasmid libraries termed "T.spiezzo" (gene ablation, 19,936 plasmids) and "T.gonfio" (gene activation and epigenetic silencing, 22,442 plasmids). The libraries and the related methodologies are described in our paper in Nature Biomedical Engineering.
These tools allow for the individual manipulation of every human gene, are broadly applicable to all human biology, and are attracting strong interest from biological and medical researchers worldwide. However, while these plasmid collections are suitable to studying immortalized transfectable cell lines, their most transformative utilization involves primary human cells and human induced pluripotent stem cells, which require that plasmid libraries be repackaged into lentiviral vectors. With CRISPR4ALL we enable the access of the global research community to these toolsets and to the research data generated therefrom by 1) generating lentiviruses from our library plasmids and providing them to the research community; 2) by providing practical training, protocols and standards to interested users, and 3) by creating a community of users adopting robust FAIR-compliant computational tools for collaborative research and data sharing.
Our libraries use quadruple-guide designs (4 sgRNAs per plasmid), engineered to tolerate human DNA polymorphisms, ensuring reproducibility across diverse genetic backgrounds.
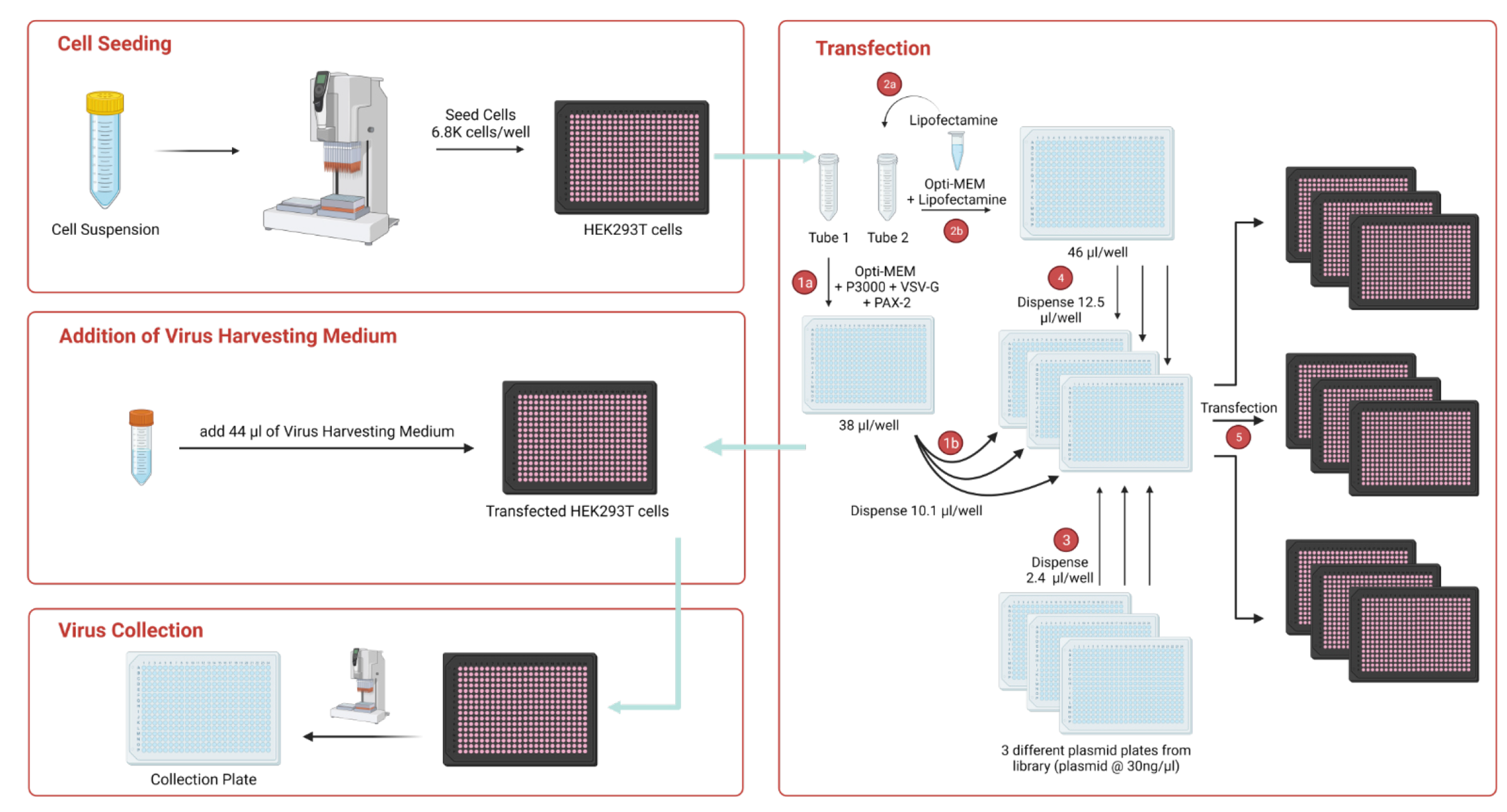
We developed Alpacloning (Automated liquid-phase Plasmid Assembly and Cloning), which allowed cloning of more than 42,000 plasmids without colony picking.
The T.spiezzo (CRISPRo) and T.gonfio (CRISPRa) sub-libraries together cover 42,146 genes, enabling truly unbiased genome-wide interrogation.
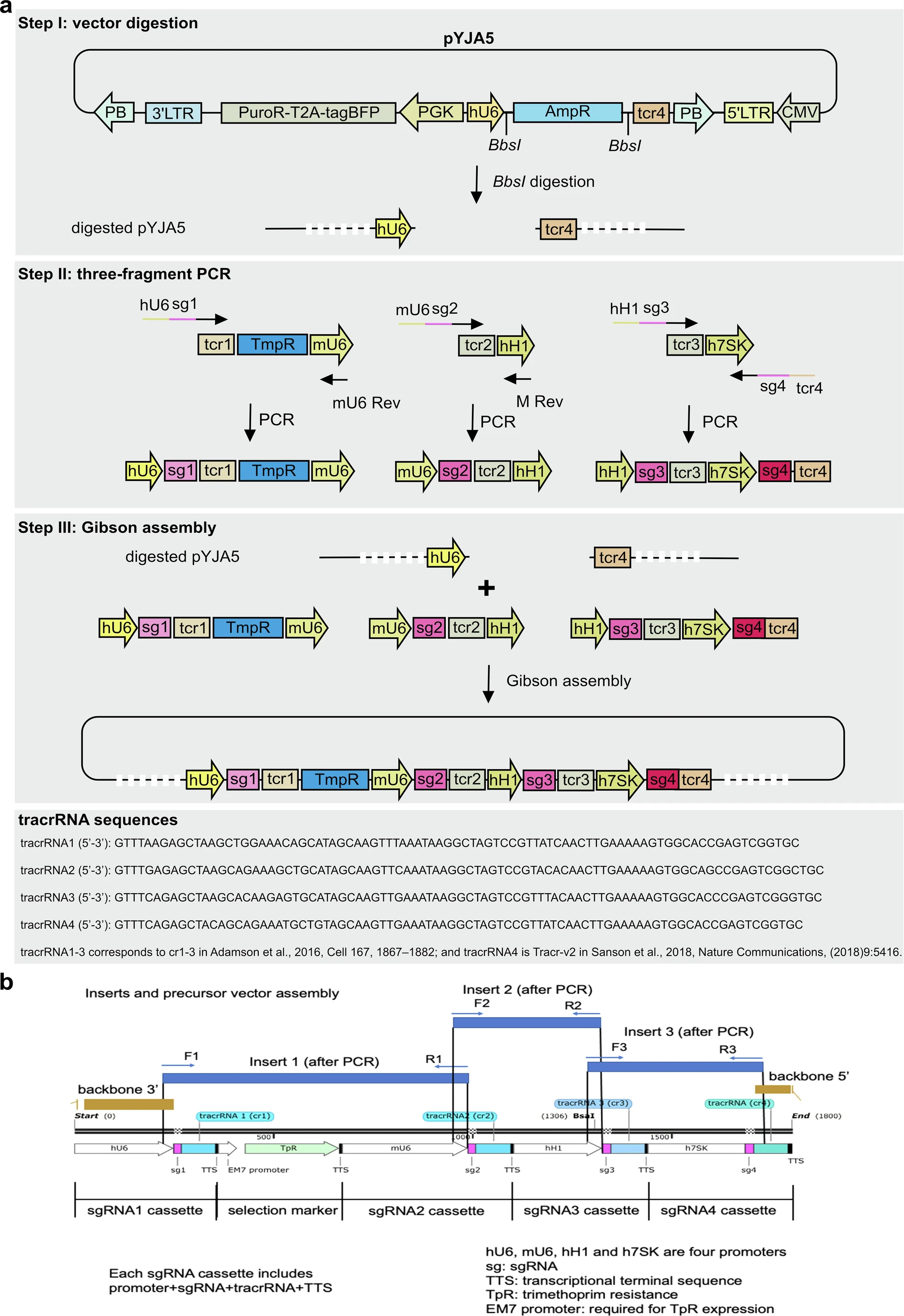
a: Step-by-step construction of qgRNA plasmids using ALPA cloning method and sequence information of tracrRNA1-4 (tcr1-4). b: Zoom-in illustration of homologous ends overlapping among the three amplicons and the digested vector pYJA5
Lentiviral production is standardized, enabling 2,000-4,000 viruses per week at high, homogeneous titers. Each lentivirus is titrated, allowing perfect normalization of pooled screens.
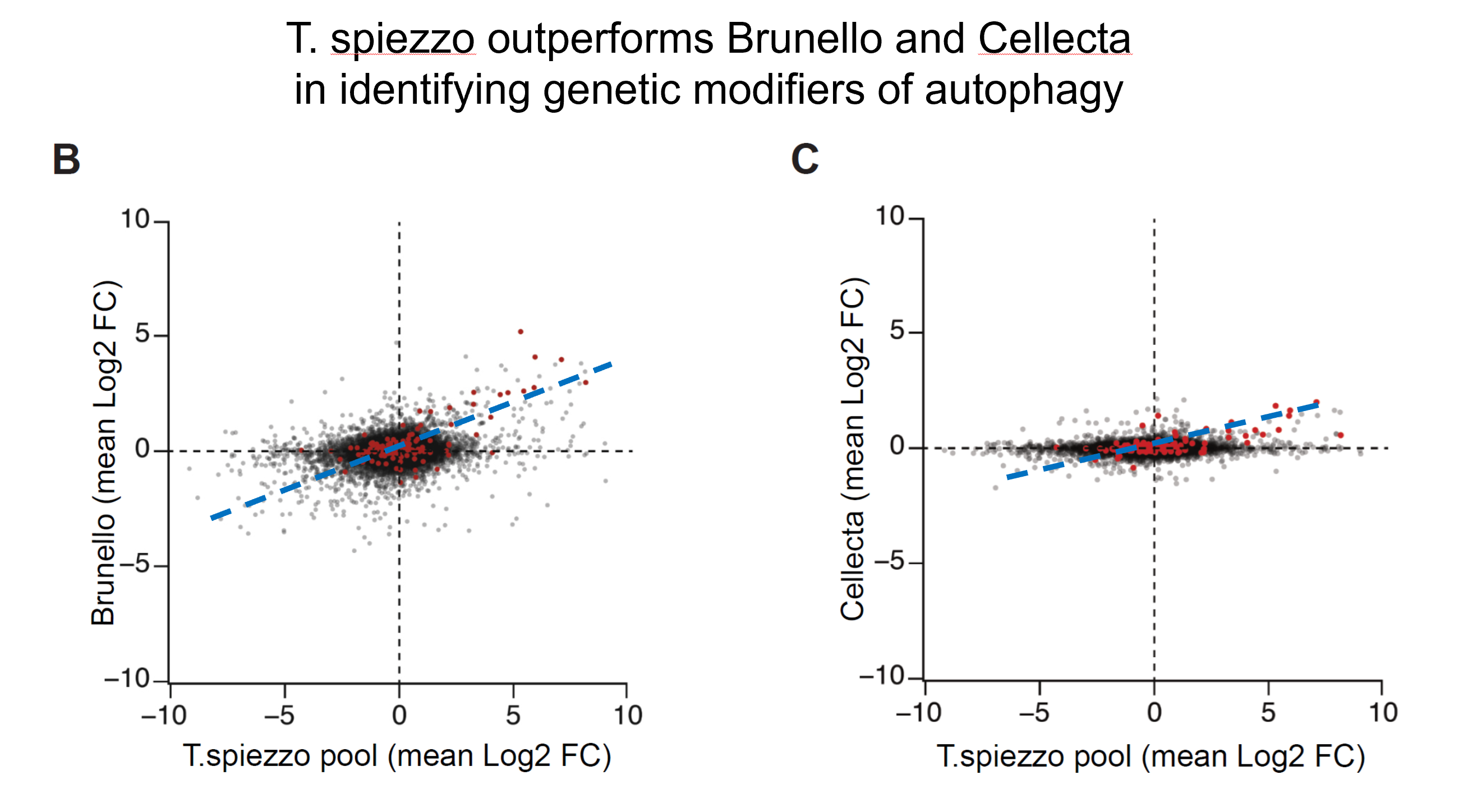
Compared to standard tools (e.g., Brunello, Cellecta), our quadruple-guide vectors consistently outperform single-guide vectors in identifying modifiers of autophagy.
These resources open the door to new territories of unbiased discovery.